Patent Sequence Search: You’re Doing It Wrong
Patent Sequence Search: You’re Doing It Wrong
Patent Sequence Search: You’re Doing It Wrong
Jun 14, 2017
 Henk Heus | Business Leader, Aptean GQ Life Sciences
Henk Heus | Business Leader, Aptean GQ Life Sciences
After 15 years in this field and writing about this many times, I’m still shocked when I see professional IP people that use BLAST for their sequence searches. BLAST is a crude and unreliable way to align sequences, and under normal circumstances it shouldn’t be used for anything patent related. There, I said it.
Sequences play such a central role in the business models of many life science companies. Therefore, I just don’t get that people spend good money on a commercial database like STN, GeneSeq or SequenceBase and are still stuck with BLAST, or something similarly flawed such as Smith & Waterman or FASTA, as their only real search option. So why is BLAST is such a problem in patent sequence searching? It comes down to two major issues:
1. BLAST Asks the Wrong Question
BLAST was written to compute the evolutionary distance between two sequences, an important biological question but hardly ever relevant to patents and IP. You don’t want to know which other sequences also have a kinase domain in them. You want to know which sequences in the database have 70% or more nucleotides in common with your entire query sequence. No amount of clever result filtering on the percentage identity and alignment length is going to compensate for BLAST’s local alignment behavior (see below). Alignments will come out wrong or will simply be missed. Smith & Waterman and FASTA do this in the exact same way and are just as unsuitable as BLAST for patent searching.
2. BLAST Is Not Reproducible
To make BLAST go faster it uses lots of statistical tricks and heuristic shortcuts. This means that algorithm parameters have to be carefully tweaked by an expert for each type of search, and that results can suddenly disappear the next time the same search is done. Especially when searching with short sequences like primers, probes and CDRs, it’s extremely common to miss real, relevant-to-your-business, hits. It goes without saying that in IP you need accurate, complete and reproducible search results and that BLAST is just not the tool to do it.
This problem was recognized and solved by us back in 2002 by implementing the GenePast algorithm and publishing about it in Nature Biotech. GenePast was specifically designed for patent sequence searching and has none of the issues that BLAST, Smith & Waterman and FASTA have. It aligns the entire query sequence with the smallest possible number of differences, like mismatches and indels. It has no heuristic shortcuts and makes no decisions about the relevance of results on its own. Therefore it produces fully accurate, complete and reproducible answers to the questions patent searchers need to ask, even when the query sequences are really short.
“So it’s a lot like Needleman & Wunsch?”, I hear you say. No, GenePast isn’t a global alignment algorithm, it is a best-fit algorithm. It doesn’t try to align the whole query sequence to the whole database sequence. Instead it finds the best possible way to fit the query sequence into the database sequence, or the other way around if the database sequence is shorter.
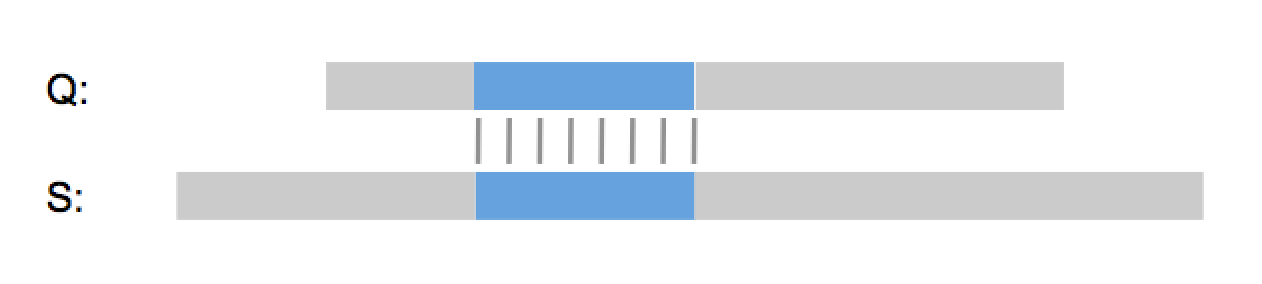
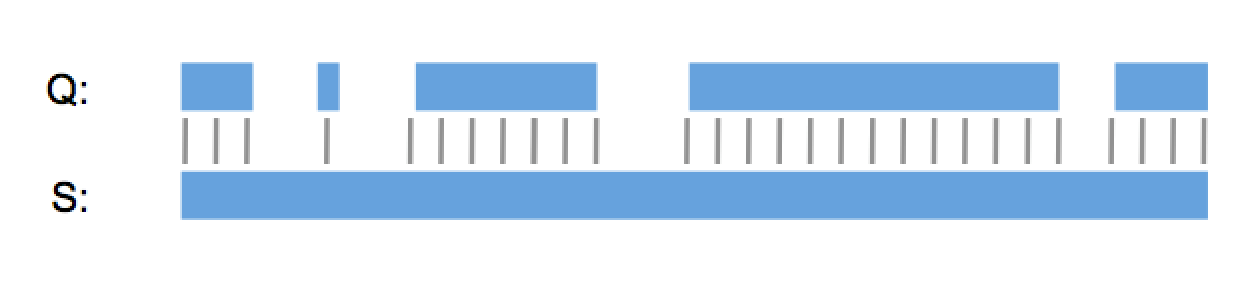
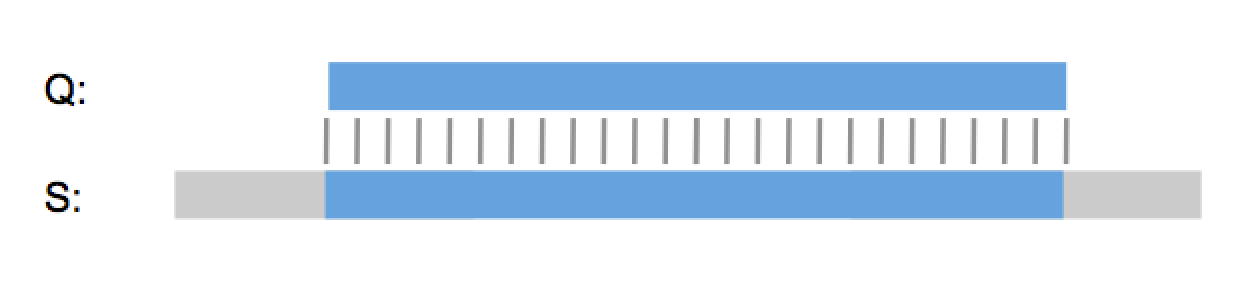
You don’t have to take my word for it. 18 out of the top 20 pharma, all five top agrochemical companies, a long list of biotech firms and law firms around the world use GenePast every day and have been for over a decade. And many major patent authorities like the EPO use GenePast for patent examination. With over 75% of the searches, GenePast is by far the most used algorithm in the Aptean GenomeQuest search interface and one of the main reasons why we’re the market leader in patent sequence searching, the other reasons being our patent sequence content of course.
With its extensive data coverage (over 500 million sequences), powerful search tools and user-friendly functionality, Aptean GenomeQuest is the obvious choice for searching the entire sequence domain, both patent and non-patent.
Avoid the pitfalls of using free solutions for IP sequence searching. Download our RFP template or start a free trial today!
Listo para tu selección de búsqueda de secuencias de PI
Usa nuestra plantilla gratuita de solicitud de propuesta (RFP por sus siglas en inglés) para identificar la solución de búsqueda de secuencia de PI adecuada para tu empresa.